JGI Seed Corn Funded Project Blog
Physical activity (PA) is among the most important human behaviours to improve and maintain health. The level of PA performed by an individual is often measured by accelerometers (the sensors used in fitness trackers or smartphones), but the obtained data is rich and evokes statistical challenges. Hence, novel statistical solutions must be found. Multivariate Pattern Analysis (MPA) could help in this regard and has great potential to provide new insights into how PA relates to health. In this first part of our 2-part blog series we describe how we will study the multivariate PA intensity signature related to early adult physical and mental health.
The problem in a nutshell
In research, accelerometers are typically worn around the hip or wrist for several days. They measure movements of the body multiple times per second and thus produce a massive amount of raw data. In general, being active will increase the measured acceleration (ie, the stored values will be higher). All values collected over the week are then used for the analysis, for example, by averaging them. This average value represents the total amount of PA performed. Another option is to look at the time spent in specific intensities of PA (eg, minutes per week of lower or higher intensity). This can be done by applying so called ‘cut points’ to the measured acceleration (the stored values). For example, if the stored value is greater than 4000, we could assume this minute was of higher intensity (those cut points are usually developed in studies where the accelerometers are compared to other measurements of the intensity of PA). Thus, cut points can be used to estimate the weekly time spent in different intensities of PA.
Many previous studies investigating associations between PA and health have focused on few intensity categories (ie, sedentary, light, moderate, vigorous). Special attention has been paid to time spent in moderate-to-vigorous PA. In fact, current PA guidelines are heavily based on this evidence. The focus on broad and selected parts of the intensity spectrum has at least two problems. First, many activities will be collapsed into the same group. For example, brisk walking and playing Squash, even though their intensity can be vastly different, are included in the same category (moderate-to-vigorous PA). Secondly, we do not know enough about the relative contribution of lower-intensity PA to health (eg, light).
However, including all the intensity categories in a single statistical model (eg, Ordinary Least Squares Regression) is problematic due to the high correlation between the variables and their closed structure (ie, summing up to 24 hours when adding sleep). Therefore, novel statistical solutions are needed to overcome these challenges and to identify the relative contribution of each intensity within the full intensity spectrum. One approach is MPA, which was, among others (eg, compositional data analysis, intensity gradient) recently introduced to the field of PA epidemiology. MPA addresses the collinearity among intensity categories using latent variable modelling (Partial Least Squares Regression (PLS-R)) while allowing for the inclusion of a high-resolution dataset (full intensity spectrum). So, instead of using the above-mentioned categories (sedentary, light, moderate, vigorous) we can not only include all the categories together but also increase their resolution by increasing the number of cut points (eg, time spent in 4000-4499, 4500-4999 instead of using just ‘4000 and greater’). Thus, single cut points (eg, 4000) are becoming less important while at the same time we can study the relative contribution of specific intensities considering all others in the same statistical model.
More information about MPA can be found here
Aims of the project
Previous applications of MPA to PA research have been cross-sectional studies on physical health (eg, cardio-metabolic health) where both the exposure (PA) and outcome (health) are measured at the same time. Therefore, the role of specific PA intensities for a broad range of physical and mental health outcomes is unknown. Moreover, given the importance of adolescence for life-course health, longitudinal studies are needed to explore the role of adolescent PA on future health. This proposed project utilises data from the Avon Longitudinal Study of Parents and Children (ALSPAC) resource, the most detailed study of its kind in the world, to provide novel evidence on associations of the PA intensity spectrum in adolescence (accelerometer measurements at ages 12, 14 and 16 years) with important adult health markers (wellbeing, depression, anxiety, cardiovascular health, metabolic health, adiposity, musculoskeletal, and respiratory health, measured at 25 years). The selected health markers are shown in the Figure below.
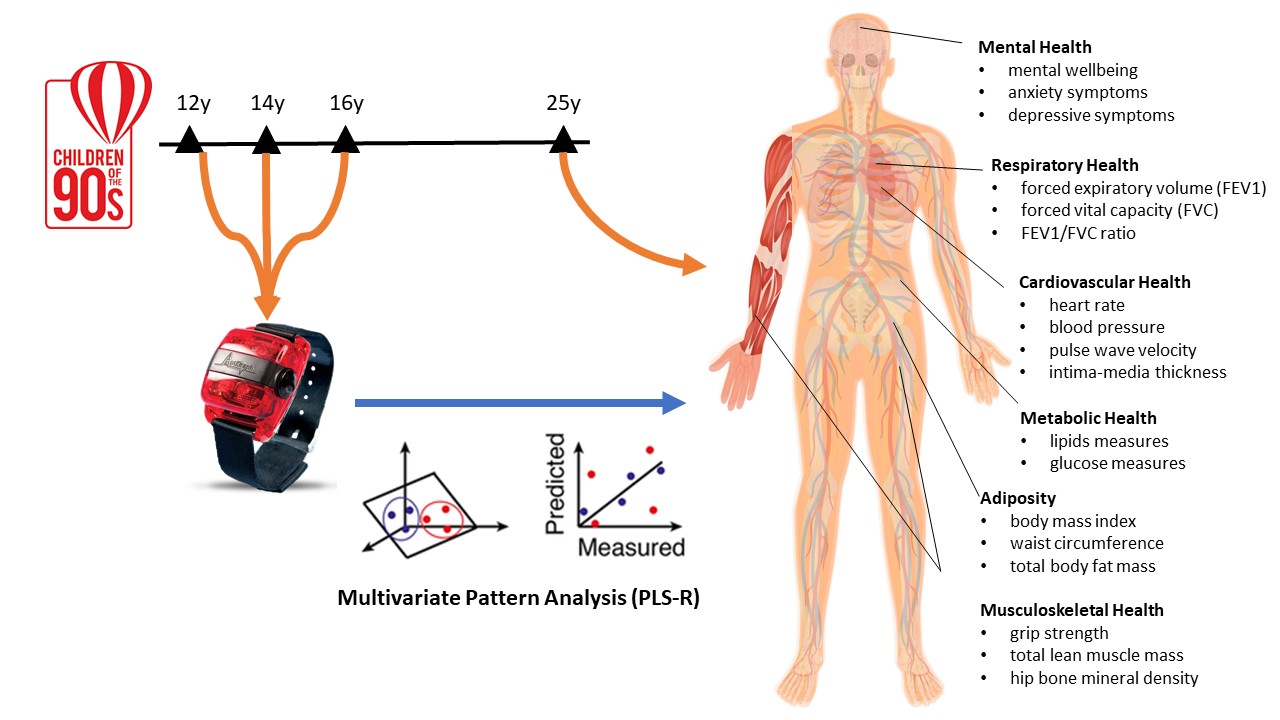
Stay tuned for Part-2 which will be published next year and shows the results of this project.
Contact details
Dr Matteo Sattler (Email: matteo.sattler@uni-graz.at, Twitter: @Sattler_Graz)
Institute of Human Movement Science, Sport and Health, University of Graz, Graz, Austria
Dr Ahmed Elhakeem (Email: a.elhakeem@bristol.ac.uk, Twitter: @aelhak19)
MRC Integrative Epidemiology Unit at the University of Bristol, Bristol, UK