JGI Seed Corn Funded Project Blog
Background
Environmental monitoring is critical for the protection of human health and the environment. As the world’s population continues to increase, industrial development and agricultural practices continue to expand, as does their associated pollution. The requirement for environmental monitoring is thus greater than ever, particularly for freshwater resources utilised for human consumption.
Biological monitoring of freshwater resources involves regular characterisation of dominant microalgal communities that are highly sensitive to nutrient pollution, forming widespread harmful algal blooms (HABs) during the process of eutrophication. But, traditional microscopy-based monitoring techniques to identify and count microalgae represent a significant bottleneck in monitoring capabilities and limit monitoring to institutions with highly trained individuals.
Project Aim
This project was founded to provide proof-of-concept for the application of artificial intelligence, specifically deep learning convolutional neural networks (CNNs), for rapid detection and identification of dominant microalgal groups and trouble HAB-forming species in freshwater samples.
Major actions
- Create robust training dataset: The first step to achieve this was to produce a robust, annotated training dataset of both controlled (i.e. mono-species cultures) and wild-type (i.e. natural) samples. A partnership with Dwr Cymru Welsh Water (DCWW) was established, allowing for the provision of water samples from their reservoirs over the spring-summer season, as well as access to their culture collections of dominant HAB-forming taxa. JGI support then allowed to recruit our intern, David Furley, who spent a month imaging and annotating images of both types of samples, with support provided from DCWW experts to ensure the highest accuracy of species identification.
Outcome 1: In total ~5000 annotated wild-type images were produced containing a variety of algal species (e.g. Figure 1), and ~3000 annotated culture-collection images, across dominant cyanobacteria, diatom and chlorophyte algal species; a major feat in such a short timeframe, well done David!
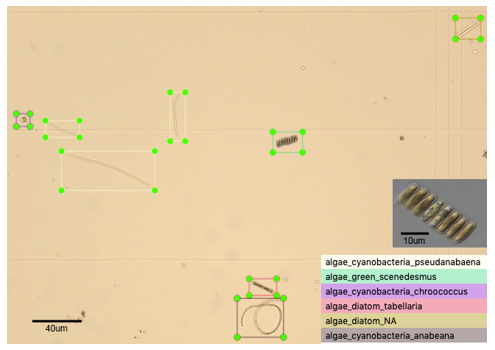
Figure 1: Representative training dataset image of microalgae found within a wild-type water sample at x100 magnification, showing bounding boxes drawn around six different genera of algae classified based on morphology and size.
- Test off-the-shelf CNNs for algal detection and identification: Once a robust training dataset was produced, the next step was to test the application of existing CNNs for the tasks of object detection (finding and drawing a bounding box around algal cells within images) and identification (assigning the correct taxonomic label to each object identified). For this proof-of-concept project, we chose to test a PyTorch implementation of the YOLO (You Only Look Once) version 3 CNN. YOLOv3 predicts bounding boxes using dimension clusters as anchor boxes, predicting an objectness score for each bounding box using logistic regression. The class each bounding box may contain is predicted using multilabel classification via independent logistic classifiers. The sum of squared error loss is used for training bounding box predictions, and binary cross-entropy loss for class predictions.
Outcome 2: YOLOv3 proved highly effective at object detection of microalgae within mono-specific culture images but more importantly, wild-type samples containing a mixture of algal species as well as non-algal particles. Overall, however, YOLOv3 performed less well at object identification.
- Test bespoke KERAS (TensorFlow) CNNs for algal identification: To build on our initial progress in algal detection, bounding boxes were used to cut algal cells from images within our training dataset, creating a second database of annotated individual algal cells to be used as input into a purely identifier focussed CNN. For this we employed a KERAS-based CNN on images comprising 3 types of algae; Oscillatoria HAB-forming cyanobacteria, Asterococcus Chlorophyte algae, and Tabellaria diatoms. Two training datasets were produced; i) a non-augmented training dataset comprising 273 images (91 from each class); and ii) an augmented training dataset that totaled ~ 6552 images (2184 from each class). Two instances of our novel CNN were then trained for 270 epochs each.
Outcome 3: Whilst the CNN trained on non-augmented images performed relatively well (Fig. 2), with identification accuracies ranging 86 – 100% across three classes of microalgae (Fig. 3), image augmentation significantly improved training outcomes, with Oscillatoria cyanobacteria identified with 97% accuracy, Tabellaria diatoms with 99% accuracy and Asterococcus green algae with 100% accuracy (Fig. 3).
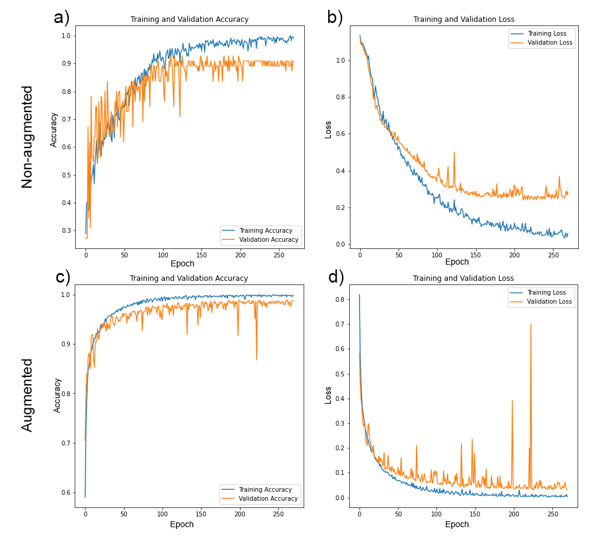
Figure 2: Training (blue lines) and validation (orange lines) accuracy (a & c) and loss (b & d) for bespoke KERAS-CNNs trained on non-augmented (a & b) and augmented (c & d) training datasets over 270 epochs.
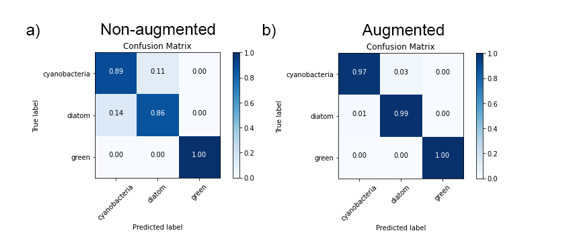
Figure 3: Confusion matrices showing classification results for validation data for our KERAS-CNNs trained on non-augmented data (a) and augmented datasets (b). Values represent percentage of correct/incorrect classifications.
Overall
This project has demonstrated proof-of-concept for the application of convolutional neural networks in the monitoring of microalgal communities within critical freshwater resources. We have amassed a sizeable, annotated training dataset of both wild-type and cultured samples, demonstrated the success of off-the-shelf CNNs in microalgal detection within images of water samples, and provided the first step on the road to developing CNNs capable of algal identification.
Future plans
Much work remains to be done on this topic before we have CNNs capable of automated algal detection, identification and enumeration from natural samples. We will continue to test different CNN architectures on our 8000 image training dataset. Collaborations with DCWW are ongoing and the outputs from this work will form the evidence base for a larger project application to drive the incorporation of CNN techniques into environmental monitoring.
Contact details:
Please contact the PI Chris Williamson at c.williamson@bristol.ac.uk and see his research group website at www.microlabbristol.org

