Blog written by Rob Arbon, Data Scientist at the University of Bristol.
This project was funded by the annual Jean Golding Institute seed corn funding scheme.
Multitask learning for AMR
“Multitask learning for AMR” developed out of our collaboration with the Jean Golding Institute on the One Health Selection and Transmission of Antimicrobial Resistance (OH-STAR) project funded by the Natural Environment Research Council (NERC), the Biotechnology and Biological Sciences Research Council (BBSRC) and the Medical Research Council (MRC).
OH-STAR is seeking to understand how human activity and interactions between the environment, animals and people lead to the selection and transmission of Antimicrobial Resistance (AMR) within and between these three sectors. As part of this project we collected thousands of observations, from over 50 dairy farms, of the prevalence of AMR bacteria, as well over 300 potential environmental variables and farm management practices which could lead to AMR transmission or selection (so called risk factors). If we can identify these risk factors the hope is that we can use this information to shape policy to reduce the spread of AMR into the human population where it threatens to cause widespread death and disease.
Multitask learning (MTL) is a statistical technique that aims to relate different “tasks” in order to improve how we perform those tasks separately and to understand how the tasks are related. MTL has been used in many different areas from improving image recognition and to help diagnose neurodegenerative diseases. In this project we wanted to see if MTL could be used to help better understand the OH-STAR data and also to sketch out ideas for potential grant applications to develop this idea further.
To evaluate MTL for our purposes, we focused our attention on two small subsets of the OH-STAR data: 600 faecal samples from pre-weaned young calves and 1800 from adult dairy cows. Each of these samples had been tested to see whether the Escherichia coli. bacteria within them contained the CTX-M gene. This gene is important because it confers resistance to a range of antibiotics such as penicillin-like and cephalosporin antibiotics which are used to treat a range of different infections in humans and cattle.
In order to model the risk of something occurring, statisticians use a technique called logistic regression. With this technique you can quantify by how much a risk factor, e.g. which antibiotics a farmer may use, increases or decreases the risk of observing the CTX-M gene in samples from the farm. As an example, consider how one of our risk factors, atmospheric temperature, affects the risk of observing CTX-M. Each dot on the chart below represents one of our samples, whether it had CTX-M (right hand axis), and the temperature when the sample was recorded (horizontal axis). The results of the logistic regression are the black line (left hand axis): the risk of observing CTX-M as the temperature increases.
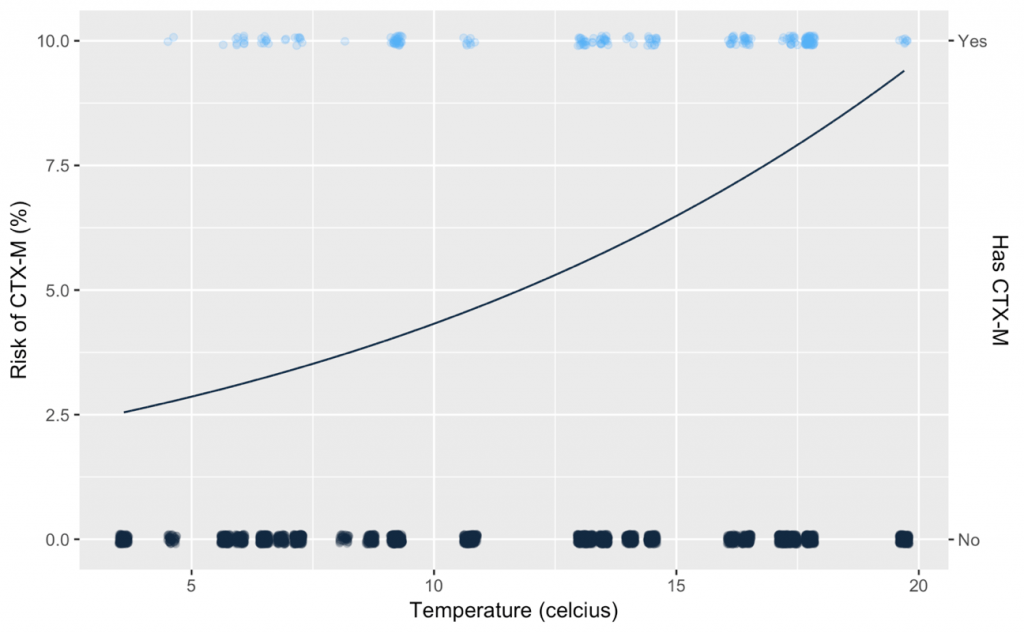
This relationship can be summarised by a single number called the log-odds-ratio, in this case the log-odds-ratio of temperature is 0.4. The fact that this number is greater than 0 means the temperature increases the risk of CTX-M, if it were less zero this means that it decreases the risk.
So, to quantify how each of the measured risk factors affects the risk of observing CTX-M we could just use logistic regression on all 300 risk factors for both the adults and separately for the calves and look at the log-odds-ratios for each risk factor. This approach suffers from two problems however:
- by fitting a standard logistic regression model with 300 risk factors and at most 1800 observations means your conclusions won’t necessarily be correct for a wider population (because of overfitting)
- this approach treats understanding risk in adults and heifers as two separate tasks, when in fact they share many similarities.
To overcome the first problem statisticians use a technique called regularization. In a nutshell this reduces the complexity of your model to prevent it predicting random points (“noise”) in your data and focuses the model on predicting the “true” signal.
Multi-task learning (MTL) is an approach to overcoming the second problem. The idea is that some risk factors will pertain more to calves than to adults (e.g. the type of antibiotics given when they are young) or vice versa. So it makes sense for these risk factors to have different impacts on the model. However, there are some risk factors that will have very similar effects on both types of animal (e.g. outside temperature ). The way MTL relates tasks is complicated but it is very similar to the regularization technique linked to above. Interested readers can read reviews of MTL in A brief review on multi-task learning and An overview of multi-task learning in deep neural networks.
We looked at a number of different MTL techniques using the R package RMTL (package and accompanying paper) but for brevity we will consider only one here. Our two ‘tasks’ were logistic regression to find the risk factors affecting pre-weaned calves (task 1, labelled ‘heifers’) and adults (task 2).
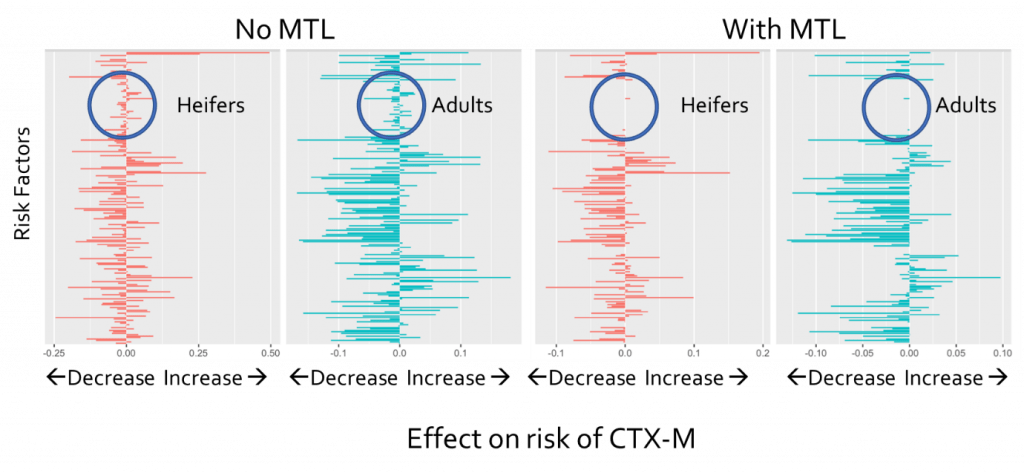
The technique I’d like to talk about here is called ‘Joint feature learning’ and relates the tasks by encouraging them to have similar values for each risk factor. This means that if they’re not important for both, they will feature less strongly in each model. The results are shown below for the results with (right) and without (left) MTL.
The red results are for heifers and the blue results are for the adults. Each bar denotes the effect of a risk factor on the risk of observing CTX-M: positive increases the risk while negative decreases the risk. The effect of using MTL was to suggest that the circled risk factors were not important to both tasks as applying MTL made all those risk factors irrelevant to the task. This was important for two reasons.
First, this cut down on the number of possible risk factors that needed further investigation. Second, this meant that those risk factors which did show up as having different effects on the risk could be trusted as not being down to just chance. For example, temperature was one of the most important features for the heifers but not for the adults – this could provide ideas for interesting hypotheses to test in future work.
MTL has potential novel applications in real world scenarios
The main conclusion from this work is that MTL has potentially novel applications in complicated “real-world” scenarios but that the tools for MTL need developing. For example the techniques in the RMTL package did not fully take into account the structure of the data. This is something that will need to be addressed in any future work.
In our wrap up meeting we discussed the potential for using these techniques in future work. The main idea discussed was to use data collected as part of a completely separate project to help understand risk in OH-STAR data and vice versa. For example, the One Health drivers of AMR in rural Thailand (OH-DART) is a similar project, funded by the Global Challenges Relief Fund (GCRF). The distinct but related datasets from OH-STAR and OH-DART projects could be analysed jointly using MTL to identify risk factors. Thanks to the funding from the JGI we now have the materials necessary to write such a proposal and we will be watching calls from e.g. the GCRF and BBSRC in the near future to fund this work.
All of our code for this work can be found on the Open Science Framework.
The Jean Golding Institute seed corn funding scheme
The JGI offer funding to a handful of small pilot projects every year in our seed corn funding scheme – our next round of funding will be launched in the Autumn of 2019. Find out more about the funding and the projects we have supported.