Hypothesis testing using statistics usually proceeds via something along these lines:
- Collect some data, say on the response of some variable in two different groups to a treatment and a control.
- State a null and alternate hypothesis. The null hypothesis is something you want to falsify and typically is a nill hypothesis i.e. one that hypothesizes no effect. In this case we might have
 . The alternate hypothesis is usually the negation of the null hypothesis.
. The alternate hypothesis is usually the negation of the null hypothesis.
- Calculate the test statistic,
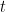 . In our case this would be the difference of the two mean responses in each group divided by the pooled variance
. In our case this would be the difference of the two mean responses in each group divided by the pooled variance 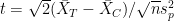 .
.
- Calculate the probability of observing
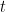 or more a extreme value under the distribution implied by the null hypothesis, this is the
or more a extreme value under the distribution implied by the null hypothesis, this is the 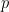 -value.
-value.
- If the
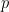 -value is less than some pre-set significance level, e.g.
-value is less than some pre-set significance level, e.g. 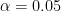 , then the we reject the null hypothesis in favour of the alternative. If
, then the we reject the null hypothesis in favour of the alternative. If  then we say we fail to reject the null.
then we say we fail to reject the null.
The hypothesis test may be on group means, on regression coefficients or some other quantity. All that is needed is the sampling distribution of the statistic in question under the null hypothesis. In our example, sample means divided by sample variance is distributed as a Student 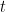 distribution.
distribution.
This process is called Null Hypothesis Significance Testing (NHST) and, n a nutshell, it is the process of accepting or rejecting a null hypothesis based on a 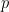 value being greater or less than a threshold (significance level).
value being greater or less than a threshold (significance level).
This post is about using NHST in your hypothesis testing, in particular this post explains:
- the history of NHST (briefly),
- why you should in most cases not use NHST,
- when you should use NHST,
- what you should do instead of NHST.
A brief history of NHST
This history and the references therein are largely a synopsis of some of the points raised in The Fisher, Neyman-Pearson Theories of Testing Hypotheses: One Theory or Two? by Lehman.
Modern (frequentist) hypothesis testing arose out of the work of Fisher and of Neyman & Pearson (N&P). The two are logically distinct but NHST has elements of both and so I’ll describe them both here.
Fisher originally proposed stating a null hypothesis and calculating 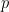 values as an index of the strength of evidence agains the null hypothesis. He suggested 5% and 1% as
values as an index of the strength of evidence agains the null hypothesis. He suggested 5% and 1% as 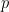 values below which count strongly against the null hypothesis (Fisher, 1946, Statistical Methods for Research Workers)
values below which count strongly against the null hypothesis (Fisher, 1946, Statistical Methods for Research Workers)
If P is between . 1 and .9, there is certainly no reason to suspect the hypothesis tested. If it is below .02, it is strongly indicated that the hypothesis fails to account for the whole of the facts. We shall not often be astray if we draw a conventional line at .05…
However he later rejected the need for standard threshold values for assessing significance (Fisher, R. A. 1956: Statistical methods and scientific inference.):
“no scientific worker has a fixed level of significance at which from year to year, and in all circumstances, he rejects [null] hypotheses; he rather give his mind to each particular case in the light of his evidence and ideas.
Fisher also believed that one should use 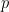 values as a method for drawing conclusions about the experimental data, rather than making decisions by accepting and rejecting hypotheses, (Fisher, 1973, Statistical Methods and Scientific Inference):
values as a method for drawing conclusions about the experimental data, rather than making decisions by accepting and rejecting hypotheses, (Fisher, 1973, Statistical Methods and Scientific Inference):
The conclusions drawn from such tests constitute the steps by which the research worker gains a better understanding of his experimental material, and of the problems which it presents.
… More recently, indeed, a considerable body of doctrine has attempted to explain, or rather to reinterpret, these tests on the basis of quite a different model, namely as means to making decisions in an acceptance procedure.
Neyman and Pearson took the the idea of cut-offs and formed a statistical decision making process. They advocated controlling type I errors (falsely rejecting the null hypothesis) by using a significance level, 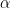 . Rejecting the null when
. Rejecting the null when 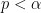 maintains the false rejecting rate at
maintains the false rejecting rate at 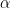 . They also suggested controlling type II errors (falsely accepting the null hypothesis) through the concept of statistical power, $\beta$. In order to do this a specific alternative hypothesis (e.g.
. They also suggested controlling type II errors (falsely accepting the null hypothesis) through the concept of statistical power, $\beta$. In order to do this a specific alternative hypothesis (e.g.  ) must be stated and sample sizes calculated to maintain a type II error rate of
) must be stated and sample sizes calculated to maintain a type II error rate of 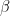 . However, N&P did advocate leaving the balancing the control of these two types of error to the experimenter, rather than having a universal cut-off.
. However, N&P did advocate leaving the balancing the control of these two types of error to the experimenter, rather than having a universal cut-off.
In summary then, both Fisher and N&P both used 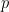 -values to test hypotheses. In the end Fisher advocated
-values to test hypotheses. In the end Fisher advocated 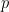 values as a continuous index of evidence against a null hypothesis and a specific set of experimental data. N&P dichotomised the
values as a continuous index of evidence against a null hypothesis and a specific set of experimental data. N&P dichotomised the 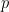 values and introduced specific alternative hypotheses in a decision making framework which attempted to control type I and type II errors.
values and introduced specific alternative hypotheses in a decision making framework which attempted to control type I and type II errors.
Over time these two approaches have blended into NHST described in the introduction. Predominantly NHST is based on the N&P paradigm. But while N&P advocated selecting significance and power levels based on scientific expediency, modern NHST has adopted the early ideas of Fisher of having levels of significance (and power) dictated by convention.
Why you (probably) shouldn’t use NHST
This section largely taken from Abandon Statistical Significance – McShane, Gal, Gelman et. al. plus some of my own thoughts. With the amount of criticism NHST has received it’s not worth summarising here so I shall just summarise those points which will have the most resonance with PhD students.
There are broadly two types of argument against NHST. The first type are those which result from features of method which are in themselves poor. The second type are those which are by-products of the method which result in undesirable outcomes. With the exception of bias in point 2 below, the criticisms presented here are of the first type. For more of both types of criticism but especially the second, see the relevant Wikipedia page.
- The null hypothesis is not (very) realistic.
- Statistical tests are only part of the evidence for or against a hypothesis.
- Failing to publish “non-signficant” results biases the literature and hurts you (less publications).
- Thresholds for deciding what’s true or not don’t make sense.
- Accepting or rejecting hypotheses is not what your publication is about.
Let’s expand on these points.
- A null hypothesis of zero effect is not realistic because of the presence of systematic error (which you may or may not be aware of) due to, amongst other things, measurement error, hidden confounders, failure to randomise etc. It’s also unrealistic because zero effects amongst the biomedical, social and clinical sciences are themselves unrealistic, e.g. no matter how you select your participants there’s unlikely to be a homogenous group which you are measuring.
- Gelman notes that
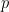 values have taken the place of discussion of other neglected factors such as prior evidence. See recommendations at the end for larger list of these factors.
values have taken the place of discussion of other neglected factors such as prior evidence. See recommendations at the end for larger list of these factors.
- Work which doesn’t attain significance goes unpublished, which is both demoralising and your publication record then fails to document your work as a researcher. In addition only publishing significant results results in a literature with biased estimates of effects. This issue is subtle and complicated and I’ll address this in another post.
- The concept of accepting or rejecting hypotheses based on a sharp threshold doesn’t make much sense when thinking about scientific hypotheses. In reality
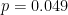 and
and  represent the same strength of evidence against the null hypothesis, but under NHST, these two results point to opposite conclusions.
represent the same strength of evidence against the null hypothesis, but under NHST, these two results point to opposite conclusions.
- A single study cannot decide on the truth or falsity of a particular hypothesis so it doesn’t make sense to frame your results that way.
When you should use NHST
Criticisms aside there are times when you might want to use NHST, such as screening and quality control purposes. For example you may want to identify genes which show an association with an effect. Typically you would consider many thousands of genes and those which do show an effect might be candidates for further research. In this case it may well be the only practical solution to have hard cut-offs for deciding which genes get studied. Another example would be quality control for industrial processes. The “hypothesis” being tested in this case is not a scientific one but one that only has bearing on whether to keep or reject a manufactured widget.
What you should do instead of NHST
The take away message of this post is to avoid, as Gelman puts it, uncertainty laundering – that is turning data into true/false pronouncements on your hypothesis. So what should you do? While there is not going to be a template applicable to all scientific areas here are some recommendations:
- You should calculate
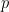 values and report their actual value, not as significant/not-significant. Realise that
values and report their actual value, not as significant/not-significant. Realise that 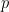 values are contingent on some very restrictive assumptions (zero effect size, zero systematic error etc.) and that a small
values are contingent on some very restrictive assumptions (zero effect size, zero systematic error etc.) and that a small 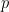 value indicates a problem with at least one of the assumptions, not just the null hypothesis.
value indicates a problem with at least one of the assumptions, not just the null hypothesis.
- Present estimated effect sizes and confidence intervals or standard errors alongside
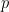 values.
values.
- Include descriptive statistics and informative visual display.
- Promote the discussion of the other aspects of the work, the neglected factors, e.g. systematic errors, prior evidence, plausibility of mechanism, study design and data collection plus any other domain specific issues.
Conclusion
At the heart of NHST is the dichotomization of 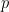 values which turns data into true/false statements about a hypothesis. This is bad practice primarily because it is both illogical and demotes other pieces of evidence (the neglected factors). Instead,
values which turns data into true/false statements about a hypothesis. This is bad practice primarily because it is both illogical and demotes other pieces of evidence (the neglected factors). Instead, 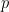 values should form only part of the statistical and non-statistical evidence for or against a hypothesis.
values should form only part of the statistical and non-statistical evidence for or against a hypothesis.
Blog piece written by Robert Arbon, Data Scientist at the Jean Golding Institute.





