The Jean Golding Institute’s Seed Corn Scheme
The Jean Golding Institute are pleased to announce the Post Graduate Researcher Seed Corn Funding awards. Every year we provide seed corn funding to Post Doctoral Researchers, but this year we are pleased to also be able to provide funding to small-scale projects for Post Graduate Researchers at the University of Bristol, which we hope will help to develop their projects further. Through our Seed Corn Funding Scheme, we aim to support initiatives to develop interdisciplinary research in data science (including Artificial Intelligence) and data-intensive research.
The Winners
Abdelwahab Kawafi is doing a PhD in Physiology working on supervised learning for computer vision mainly on CT scans and microscopy. This seed corn project is in material science, working on particle tracking of 200nm colloids using super-resolution confocal microscopy for the analysis of the glass transition.
 Ahmed Mohammed is a postdoctoral researcher in the field of water engineering. His PhD research investigates precipitation forecasting at high spatial and temporal scales using various methods such as numerical weather prediction, optical flow-based models, and deep learning techniques.
Ahmed Mohammed is a postdoctoral researcher in the field of water engineering. His PhD research investigates precipitation forecasting at high spatial and temporal scales using various methods such as numerical weather prediction, optical flow-based models, and deep learning techniques.
Ahmed and Hongbo are working together on a project called “Performance evaluation of a deep learning model in short-term radar-based precipitation nowcasting”. Radar precipitation data will be used in this project to refine an existing convolutional neural network precipitation nowcasting model in order to improve end-to-end rainfall nowcasting model performance and compare it to optical flow-based models and the Eulerian persistence baseline model.
 Hongbo Bo is a Ph.D. student in computer science. His research focuses on social network analysis by using graph data mining techniques like graph neural networks, continual learning, and contrastive learning methods.
Hongbo Bo is a Ph.D. student in computer science. His research focuses on social network analysis by using graph data mining techniques like graph neural networks, continual learning, and contrastive learning methods.
 Anuradha Kamble is a Postgraduate Research student at Bristol Composite Institute in the Department of Aerospace Engineering. Prior to this, she had three years of industrial experience in the commercial HVAC (Heating Ventilation and Air Conditioning) and EV (Electric Vehicle) industries. Recent discoveries in materials science have seen growing applications of machine learning for materials discovery by leveraging the experimental data generated over many years in different studies. Her research project is titled “Exploiting the deep learning technique to study a novel nano-modified polymer composite.” The goal of this study is to implement deep learning techniques to investigate the most suitable parameters e.g., material composition, processing temperature, and time, to study a nano-modified polymer composite. The objective of this project is to improve the generalizability of deep learning model so that it can be widely applicable to any novel materials developed in the University of Bristol. Similar applications in the field of bio sciences will be explored by collaborations with other universities across the UK.
Anuradha Kamble is a Postgraduate Research student at Bristol Composite Institute in the Department of Aerospace Engineering. Prior to this, she had three years of industrial experience in the commercial HVAC (Heating Ventilation and Air Conditioning) and EV (Electric Vehicle) industries. Recent discoveries in materials science have seen growing applications of machine learning for materials discovery by leveraging the experimental data generated over many years in different studies. Her research project is titled “Exploiting the deep learning technique to study a novel nano-modified polymer composite.” The goal of this study is to implement deep learning techniques to investigate the most suitable parameters e.g., material composition, processing temperature, and time, to study a nano-modified polymer composite. The objective of this project is to improve the generalizability of deep learning model so that it can be widely applicable to any novel materials developed in the University of Bristol. Similar applications in the field of bio sciences will be explored by collaborations with other universities across the UK.

Tian Li is a PhD student studying Physical Geography in the School of Geographical Sciences. Her work focuses on using Earth Observation techniques to study the polar glaciers. Tian’s Seed Corn project is “An automated deep learning pipeline for mapping Antarctic grounding zone from ICESat-2 laser altimeter”. This study will pioneer the application of deep learning to satellite altimetry in mapping the Antarctic grounding zone, which is a key indicator of the ice sheet instability. Through this project, Tian aims to develop a novel deep learning framework for mapping different grounding zone features by training the NASA ICESat-2 laser altimetry dataset. This research will contribute to a more efficient and accurate evaluation of grounding line migration, with which we can better understand the contribution of Antarctica to future sea-level rise.
 Levke Ortlieb is a final year PhD student in Physics. She works on supercooled liquids and glasses. Levke uses 200nm colloids as a model system in 3D using super-resolution confocal microscopy. Detecting the particle positions is very challenging so she and Abdelwahab Kawafi are using convolutional neural networks for particle tracking.
Levke Ortlieb is a final year PhD student in Physics. She works on supercooled liquids and glasses. Levke uses 200nm colloids as a model system in 3D using super-resolution confocal microscopy. Detecting the particle positions is very challenging so she and Abdelwahab Kawafi are using convolutional neural networks for particle tracking.
More information
For more information about other funding we have provided and schemes we offer, find out more on our Funding page, and take a look at previous projects we have supported, on our Projects page.


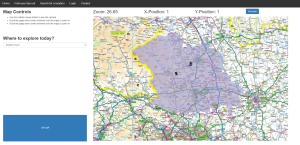
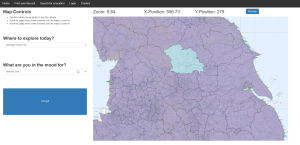
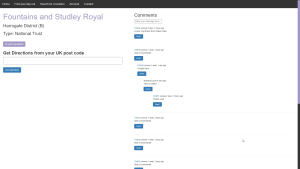 The result is a functional, although not particularly optimised solution, for exploring the 100,000’s of data points within OS Open Greenspace [4]. As an extension, I also generalised the database loader to allow for external open-source locations, such as from the National Trust [5] of English Heritage [6], to also be explored. Users can search for locations by name or narrow down what type of location and where it is, by using place location filters and the OS Boundary data [7]. Once they find a location, they can also get directions from their own address via linking to google maps. The final addition was adding some basic social media type functions, such comments and favourites, to make having a user account mean something, rather than just an authentication system for the sake of having one.
The result is a functional, although not particularly optimised solution, for exploring the 100,000’s of data points within OS Open Greenspace [4]. As an extension, I also generalised the database loader to allow for external open-source locations, such as from the National Trust [5] of English Heritage [6], to also be explored. Users can search for locations by name or narrow down what type of location and where it is, by using place location filters and the OS Boundary data [7]. Once they find a location, they can also get directions from their own address via linking to google maps. The final addition was adding some basic social media type functions, such comments and favourites, to make having a user account mean something, rather than just an authentication system for the sake of having one.  What did you enjoy about the internship?
What did you enjoy about the internship? An Interview with JGI Intern, Debby Olowu
An Interview with JGI Intern, Debby Olowu