Introduction
This project aims to investigate the role of medical experts on Twitter in influencing public health discourse and practice in the fight against COVID-19.
Aims of the Project
The project focuses on medical experts as the driving force of Network of Practices (NoPs) on social media and investigates the influence of these networks on public health discourse and practice in the fight against COVID-19. NoPs are networks of people who share practices and mobilise knowledge on a large scale thanks to digital technologies. We have chosen Twitter as a focus of our analysis since it is an open platform widely used by UK and international medical experts to reach out to the public. A key methodological challenge that this project seeks to address is to extend existing text analytics and visual network analysis methods to identify latent topics that are representative of practices and construct multimodal networks that include topics/practices and actors as nodes and Twitter affordances as edges of the network (e.g. retweets, @metions). To address this challenge, the aims of this project are:
- Build a machine learning classifier of tweets that mention relevant practices in the fight against COVID-19.
- Build a machine learning classifier of authors of tweets, which can distinguish between medical experts and other key actors (e.g. public health organisations, journalists).
Results
1. Data Collection
We used the report from Onalytica to identify the top-100 influential medical experts on Twitter. After receiving academic access to Twitter API, we collected a total of 424,629 tweets from the official accounts of these medical experts with the R package academictwitteR from 01 December 2020 to 02 February 2022.
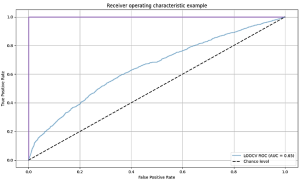
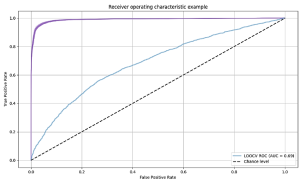
2. Build a machine learning classifier for relevant practices
After cleaning the data set, we randomly selected a sample of 1,200 tweets, which was then manually coded as either “relevant” or “non-relevant” by two independent coders and employed to train the Machine Learning classifier. By relevant we mean representative of relevant practices in the fight against COVID-19 (e.g., wearing a mask, getting a vaccine). After training and testing a series of algorithms (support vector classifier, random forest, logistic regression and naïve Bayes), a support vector classifier (SVC) gave the best classification results with 0.907 accuracy. To create the inputs to the classifier, we used a sentence transformer to convert each tweet to feature vector (a sentence embedding) that aims to represent the semantics of the tweet (https://www.sbert.net/). We compared this to a feature vector representing the number of occurrences of individual words in the tweet, achieving lower performance of 0.873 with a random forest classifier. For reference, the baseline accuracy when labelling all classes as relevant is 0.57, showing that the classifier can learn a lot from simple word features. The performance of SVC, random forest and logistic regression was similar throughout our experiments, suggesting that the choice of classifier itself is less important than choosing suitable features to represent each tweet. We applied the chosen SVC + sentence embeddings classifier to the remaining sample of tweets, resulting in 235,320 tweets that were classified as representative of relevant practices.
3. Topic modelling
We employed a topic modelling analysis to gain a better understanding of the types of practices that were discussed by the medical experts. After testing a number of indexes, we found 20 latent topics, were present in our data. We therefore employed a LDA (Latent Dirichlet Allocation) topic model analysis with 20 topics (Figure 1).

We selected 9 topics related to significant practices linked to the fight against Covid-19:
- Topic 1 is about vaccines
- Topics 2 and 16 are about global health policy/practices
- Topic 6 is about prevention of long covid in children
- Topic 9 is about immunity (either natural or vaccine-induced) against variants, hence related to COVID-19 public health measures or practices
- Topic 13 is about reporting of COVID cases and therefore linked to effectiveness of public health measures or practices
- Topic 18 is about masks (in schools)
- Topic 19 is about testing
- Topic 20 is not about a “public health” practice but a scientific practice about sequencing
4. Build a machine learning classifer of authors of tweets
One hundred twitter bios of medical experts were not enough to build the machine learning classifier. Therefore, we upsized our sample by including the bio descriptions of accounts that medical experts followed on Twitter. This strategy allowed us to include bios of users who were not medical experts and therefore differentiate between medical and non-medical “experts” or influencers. We collected the “following” accounts with the R package “twitteR” resulting into a total of 315,589 bios. We randomly selected a smaller sample for the manual coding of these bios (2,000). Following an inductive approach, two independent coders manually coded the bios into labels that classified individuals by their job occupation/profession and organizations by their sector or mission. The label “non classifiable” was used for bios that could not be classified in any professional or organizational category. This resulted into a total of 188 labels which were then aggregated into higher-level categories resulting into a final list of 49 labels.
Future plans for the project
We will use the coded sample of Twitter bios to train a Machine Learning classifier of authors of tweets. We will apply for further funding to improve our methodology and extend the scope of our project, for example, by including more medical conditions, non-English speaking countries, and other platforms in addition to Twitter. We will improve the methodology by identifying experts from a sample of collected Tweets by relevant topics representative of practices and sample and classify authors’ bios from this sample. This will allow us to have a more representative sample of individuals and organizational entities that are active in the public health discourse related to COVID-19 and other medical conditions on Twitter. We will classify practices and then map classified practices and authors onto a network to conduct a network analysis of how medical influencers affect discourse about public health practices on Twitter.
Contact details
For further information or to collaborate on this project, please contact Dr Roberta Bernardi (email: roberta.bernardi@bristol.ac.uk)